




Our next guest speaker for this semester will be, Dr. Hyun Youk, an Associate Professor of Systems Biology at UMass Chan Medical School. He received his B.S. in Physics and Mathematics from the University of Toronto, his M.A. in Astronomy and Physics from Johns Hopkins University, and his Ph.D. in Physics from Massachusetts Institute of Technology. His lab began in January 2015 at the Kavli Institute of Nanoscience in Delft, the Netherlands. After nearly six years, he moved to the University of Massachusetts Medical School (UMass Med).
His passion in life is understanding what it means to “live” and “die.” Is death inevitable? If so, why is that? If not, what are we and all other organisms doing wrong? In our attempt to address these questions, our lab seeks quantitative principles that dictate the dynamics of cellular systems. These systems include cell-signaling circuits, populations of interacting cells, and organisms that appear to be “dead” but are, in fact, merely at the nexus of life and death (e.g., dormant yeast spores).
Abstract:
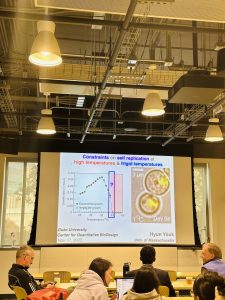
One of the hallmarks of a living cell is that it can replicate itself. An important question is when and why a cell might permanently lose its ability to proliferate and thereby transition into being a dead cell. The design principles that govern such “life-to-death” transition remain incompletely understood. In this talk, he will describe two experimental studies in which we used the budding yeast, S. cerevisiae, to reveal such design principles in the context of high and frigid temperatures. Temperature is a universal parameter for life in that it controls the speed of all biochemical reactions in all organisms and every habitat.
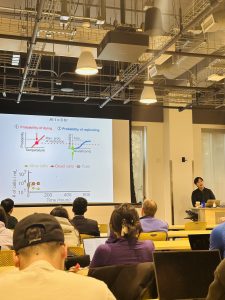
By either increasing the temperature to sufficiently high values or decreasing the temperature to sufficiently low values, we placed yeast cells at the (apparent) edge of their capacity to duplicate. For both high temperatures (> 38 C) and near-freezing temperatures (0 C – 5 C), we found ways to extend yeast’s ability to duplicate: we could enable more cells to duplicate with drastically shortened doubling times. We constructed “phase diagrams” that describe cell-population growths for both temperature regimes.
The same mathematical model, with one free parameter, reproduced both phase diagrams as well as stochastic proliferation of individual cells. At near-freezing temperatures, we discovered “speed limits” – slowest and fastest possible doubling times – at which a yeast cell’s life can progress: a cell that progresses more slowly than a “low-speed limit” defined for each temperature faces a certain death. A mathematical model and experimental data elucidated how these speed limits arise. These findings establish a quantitative foundation for engineering organisms that can survive extreme temperatures and elucidating fundamental limits to slowing down life.
ICYMI: Dr. Adam Gormley, Associate Professor of Biomedical Engineering at Rutgers University, presented his research to our seminar class regarding “Automation and Active Learning for the Autonomous Design of Polymer Biomaterials” and the nuances of drug delivery and tissue engineering that takes place in the Gormley Lab.





Our next guest speaker will be Dr. Adam Gormley on Friday, November 3, 2023 at 3:00 pm in Wilkinson 132. Dr. Gormley is an Associate Professor of Biomedical Engineering at Rutgers University and an expert in nanobiomaterials. Prior to Rutgers, Adam was a Marie Skłodowska-Curie Research Fellow at the Karolinska Institutet (2016) and a Whitaker International Scholar at Imperial College London (2012-2015) in the laboratory of Professor Molly Stevens. He obtained his PhD in Bioengineering from the University of Utah in the laboratory of Professor Hamid Ghandehari (2012), and a BS in Mechanical Engineering from Lehigh University (2006). In January 2017, Adam started the Gormley Lab which seeks to develop bioactive nanobiomaterials using robotics and artificial intelligence. Dr. Gormley is currently the PI of an NIH R35 MIRA Award, an NSF CBET Award, and an NSF Designing Materials to Revolutionize and Engineer our Future (DMREF) Award. He was recently named a Rising Star by Advanced Healthcare Materials, is the recipient of the A. Walter Tyson Assistant Professorship, the Young Innovator Award by Cellular and Molecular Bioengineering, and the Presidential Fellowship for Teaching Excellence.
Gormley’s abstract: The seamless integration of synthetic materials with biological systems long remains a grand challenge, often curtailed by the sheer complexity of the cell-material interface. For decades, biomaterial scientists and engineers have designed around this complexity by rationally designing new materials one experiment at a time. However, recent advances in laboratory automation, high throughput analytics, and artificial intelligence / machine learning (AI/ML) now provide a unique opportunity to fully automate the design process. In this seminar, we put forth our efforts to develop a biomaterials acceleration platform (BioMAP) (i.e., self-driving biomaterials lab) that can rapidly iterate through design spaces and identify unique material properties that perfectly synergize with biological complexity.
ICYMI: We would like thank Dr. Sarah Shelton, Assistant Professor of Biomedical Engineering- UNC-NCSU for discussing with our class her keen research and areas of expertise surrounding “Vascular Microphysiological Systems for Modeling the Tumor Microenvironment”.



ICYMI: On behalf of the Duke Center for Quantitative Biodesign, we thank all attendees, invited speakers, faculty, and staff for making the 2nd Annual Biomolecular Condensates Symposium during Fall Break a success. Your expertise and participation are greatly appreciated. We are already looking forward to next year’s symposium.




The Duke Center for Quantitative Biodesign mourns the loss of our esteemed colleague, Dr. Philip Benfey, an investigator of the Howard Hughes Medical Institute and the Paul Kramer Professor of Biology. Dr. Benfey was a distinguished scientist, known for his pioneering work in genetics, molecular biology, and mathematical modeling. His groundbreaking research on cellular identity and root development in Arabidopsis thaliana was truly transformative.
Dr. Benfey’s ability to bridge theory and practice was exemplified by his founding of three companies—GrassRoots Biotechnology, Hi Fidelity Technologies, and Ground Control Robotics. He was recognized as a fellow of the American Association for the Advancement of Science and a member of the US National Academy of Sciences.
Beyond his scientific achievements, Dr. Benfey was a warm-hearted mentor and friend who inspired all who knew him. His legacy will continue to inspire future generations of scientists.
Our deepest condolences to Dr. Benfey’s family, students, friends, and colleagues. His memory will forever guide us in our pursuit of scientific excellence.

Biography:
Dr. Shelton earned her B.S. and M.S. degrees in Environmental Sciences and Engineering at the University of North Carolina before joining the UNC-NCSU Joint Department of Biomedical Engineering for her Ph.D. During her doctoral studies under the guidance of Paul Dayton, she developed ultrasound contrast imaging and analysis methods to identify tortuous vasculature for improved cancer diagnosis. She earned a F99K00 award from the NIH to continue her studies of the vascular tumor microenvironment through a postdoctoral fellowship at Massachusetts Institute of Technology with Roger Kamm, with a co-appointment at Dana-Farber Cancer Institute with David Barbie. She rejoined the UNC-NCSU Joint Department of Biomedical Engineering as an Assistant Professor in 2023, and her current research is in the development of microfluidic, organ-on-chip models of disease to uncover how the tissue microenvironment and cellular interactions shape pathology and treatment response.
Abstract:
Microphysiological systems or “organ-on-chip” devices are three-dimensional models of simplified biological tissues that have expanded the types of hypotheses that can be explored in vitro. My work focuses on vascularized models of the tumor microenvironment to understand how the endothelial barrier interacts with circulating cells and the surrounding stroma in order to investigate factors that drive growth, metastasis, and resistance to therapy in oncology.One illustration of the capabilities of these models is the observation of metastasis-on-chip. By perfusing cancer cells through microfluidic vascular models in the presence of plasma proteins, we have begun to uncover how the clotting cascade influences the extravasation of cancer cells. Additionally, I developed vascularized models of the tumor microenvironment using cells from surgical resections to generate patient-specific devices. In this model, cancer-associated fibroblasts altered several functional indicators of endothelial phenotype including vascular morphology, barrier function, angiogenesis, and immune cell recruitment, likely through cytokine signaling. These types of devices allow us to dissect cellular interactions that drive disease using real-time imaging and other biological techniques.

Join us at the 2nd Annual Bimolecular Condensates Symposium at Duke University on both October 16th-17th 2023.
For questions or information, contact Xavier Larkin at xavier.larkin@duke.edu
Please see the attached schedule below:Biomolecular Condensates Symposium- Final Schedule

Biography:
Dr. Clay Wright’s research aims to understand how signaling networks facilitate both plasticity and robustness in plant form and function and to harness this knowledge to engineer proteins, signaling networks, and biosynthetic pathways for applications in agriculture and biotechnology. He received a B.S. in Chemical and Biomolecular Engineering from North Carolina State University prior to his Ph.D. in Chemical and Biomolecular Engineering from Johns Hopkins University and a Postdoctoral Fellowship in the Departments of Biology and Electrical Engineering at the University of Washington. Clay joined Virginia Tech as Assistant Professor in the department of Biological Systems Engineering. The Wright Plant Synthetic Biology lab integrates approaches from synthetic and computational biology, protein engineering, bioinformatics, molecular evolution, and genetics to quantify signaling dynamics, genetic interactions, and functional relationships in plant signaling.
Abstract:
Humanity is faced with an enormous challenge in the coming decades. The world’s population is rapidly growing, and we need to produce enough food, fuel, medicine and goods to support this growth in an environmentally sustainable and restorative way. Plants will inevitably provide many solutions to the problems we face, but we need to build environmentally sustainable, carbon-negative industries as soon as possible. To accelerate the development of improved agricultural systems that can produce more while using less, we apply synthetic biology approaches to map sequence-function relationships in plant signaling pathways and reengineer them. Towards this end we have developed genetically encoded, ratiometric biosensors for the plant growth hormone auxin in the model yeast Saccharomyces cerevisiae to reengineer how plants respond to this critical hormone. These biosensors have improved quantitative functional studies as well as directed evolution of plant auxin perception machinery. Additionally, these sensors can measure the production of auxin during different growth conditions and phases for S. cerevisiae, and may help us better understand auxin as an interkingdom signaling molecule. To effectively scale our reengineering efforts and expedite expansions to other signaling pathways we have recently developed open-source software for building plasmid and strain libraries using low-cost robotics.